Open Source Data Engineering with Front End featured in Harvard Medical School
About the project
Challenge
With Genomic Data becoming increasingly in need for privacy consideration, there's never been a more pressing time to have the data that matters most to us be securely searchable from the comfort of one's own computer. With a multitude of sites with unknown futures attempting to grab your genetic data, why not be able to search for your genomic data carefully from within your home computer.
Our solution
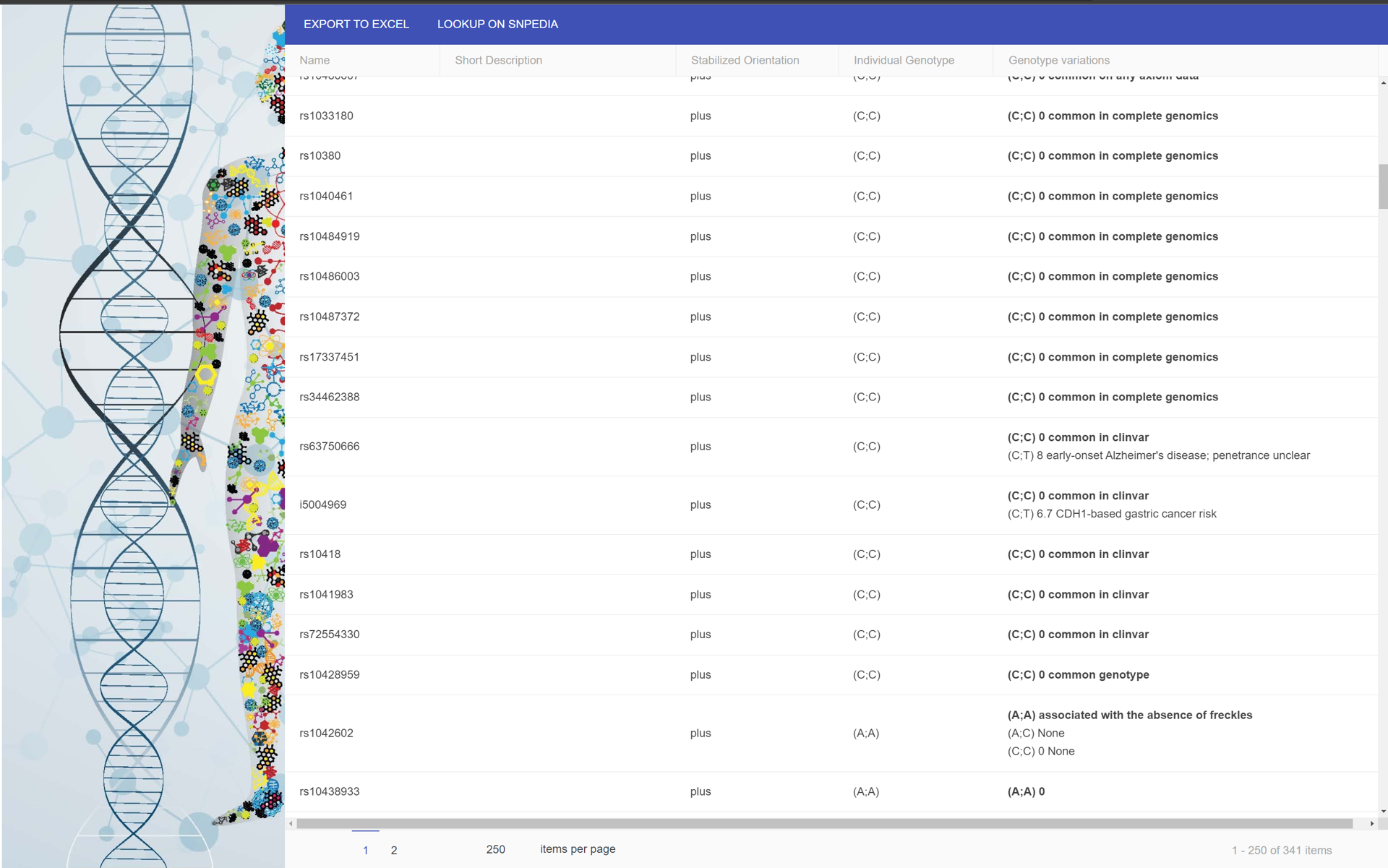
We aimed to structure the information and compile a detailed app flow that considered all the functionality and relationships in the 23AndMe product. Enter OS Genome. OS Genome allows you to examine all your genetic makeup provided by 23AndMe raw export and push to a responsive grid, where you can search 20,000 SNP's that might be meaningful to you. The application works like an archaeological dig site, gradually discovering more entries as it maps to your raw data.
Visual style
Team reached a straightforward and logical design owing to a visual style guide we created. We tackled UI and UX issues by unifying all aspects of the product into a single system with a consistent style. As a Single Page Application with Kendo Grid. The site can allow rich responsive layers from the comfort of a API endpoint. Hosted locally, this page is quick to load and quick to perform massive data visualization.
Implemented features
OSGenome is a local deployment of a responsive grid built on Kendo Grid and Python's Flask Library
01.
Kendo Responsive Grid
Kendo Responsive Grid is a Responsive JS Library that handles things quickly and easily
Conclusion
Results
We created a simple and effective page structure that has been favorited over 120+ times and is featured in Harvard Medical School's BioGrid.
Have an idea?